DCARS
DCARS.Rdthe DCARS function
DCARS(dat, xname, yname, W = NULL, rangeMin = 0, wcormin = 0, statmin = 0, extractTestStatisticOnly = FALSE, extractWcorSequenceOnly = FALSE, plot = FALSE, niter = 100, extractPermutationTestStatistics = FALSE, verbose = FALSE, ...)
Arguments
| dat | a genes x samples gene expression rank matrix, should be already converted to ranks with first column lowest survival and last column highest survival |
|---|---|
| xname | name of row of dat to test together with yname |
| yname | name of row of dat to test together with xname |
| W | weight matrix for weighted correlations, |
| rangeMin | minimum range of weighted correlation vector to include for permutation testing |
| wcormin | minimum absolute value weighted correlation vector to include for permutation testing |
| statmin | minimum value DCARS test statistic to include for permutation testing |
| extractTestStatisticOnly | if TRUE, extract only the DCARS test statistic without permutation testing |
| plot | if TRUE plot observed weighted correlatin vector |
| niter | number of iterations for permutation testing |
| extractPermutationTestStatistics | if TRUE, extract only the DCARS test statistic without permutation testing |
| verbose | if TRUE, print updates |
| ... | additional arguments passing on to weightMatrix() |
| extractWcorSequence | if TRUE, extract only the weighted correlation vector without permutation testing |
Value
either single value (p-value or test statistic), vector (local weighted correlation), or list (combination of above) depending on the input parameters
Examples
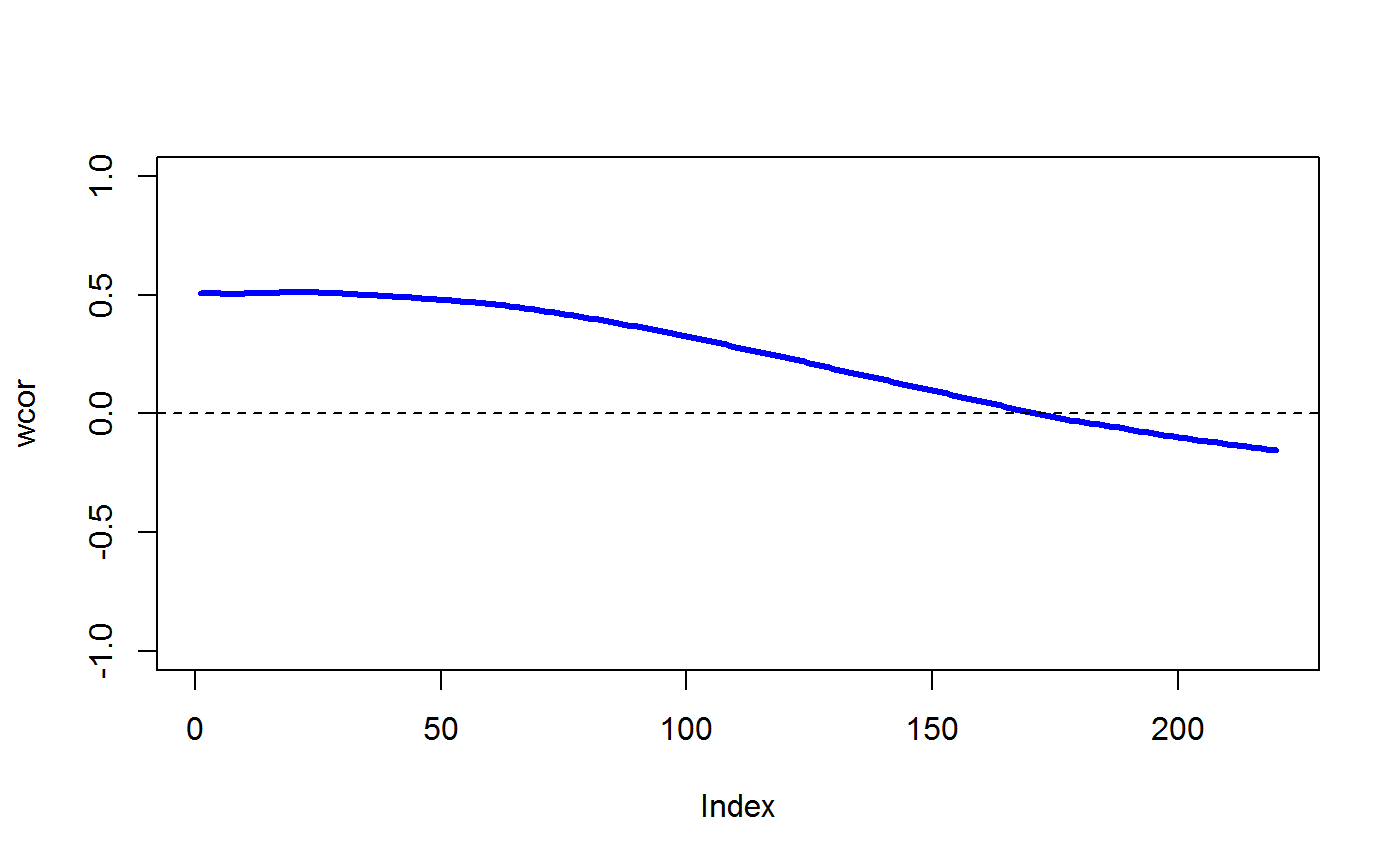
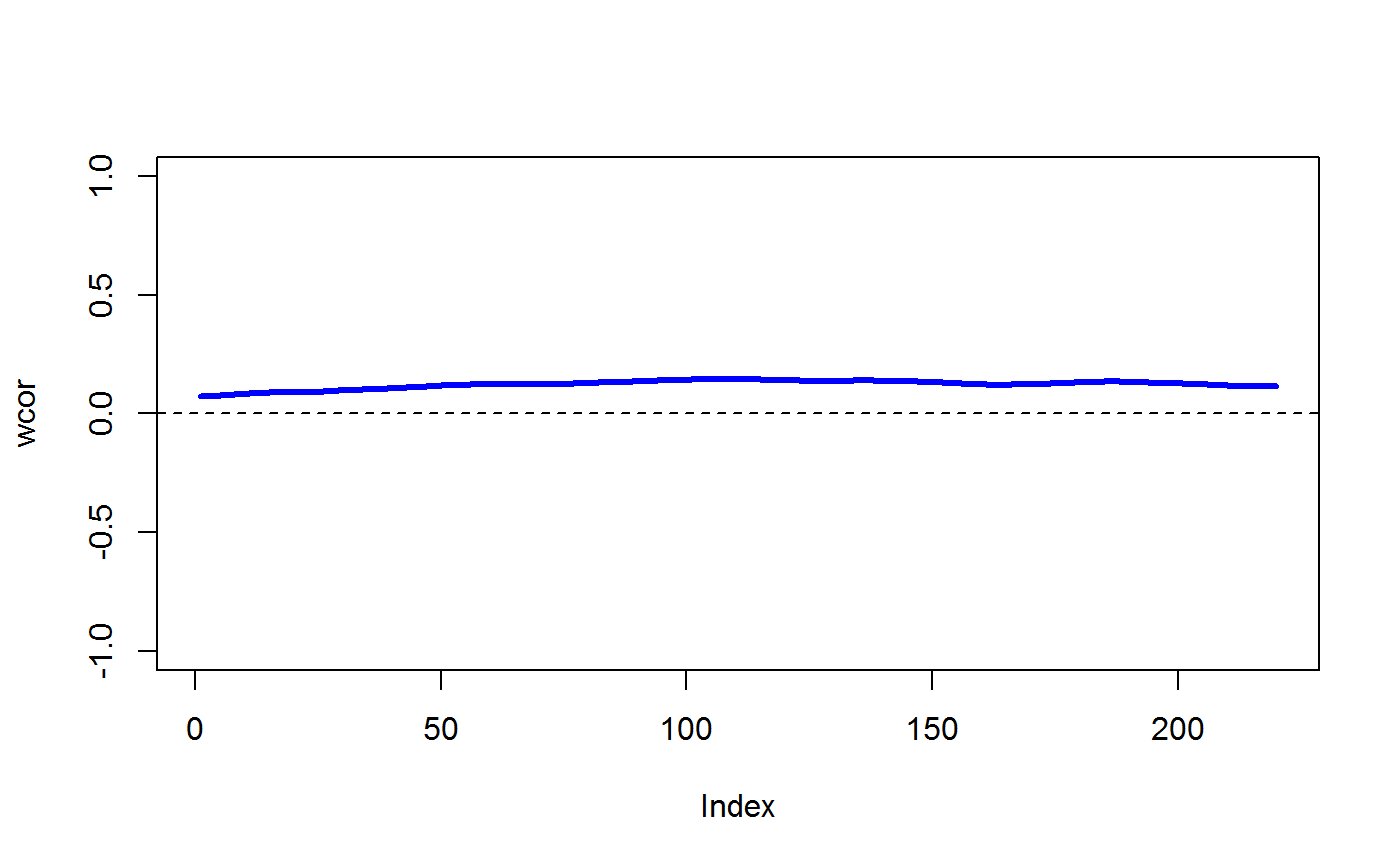
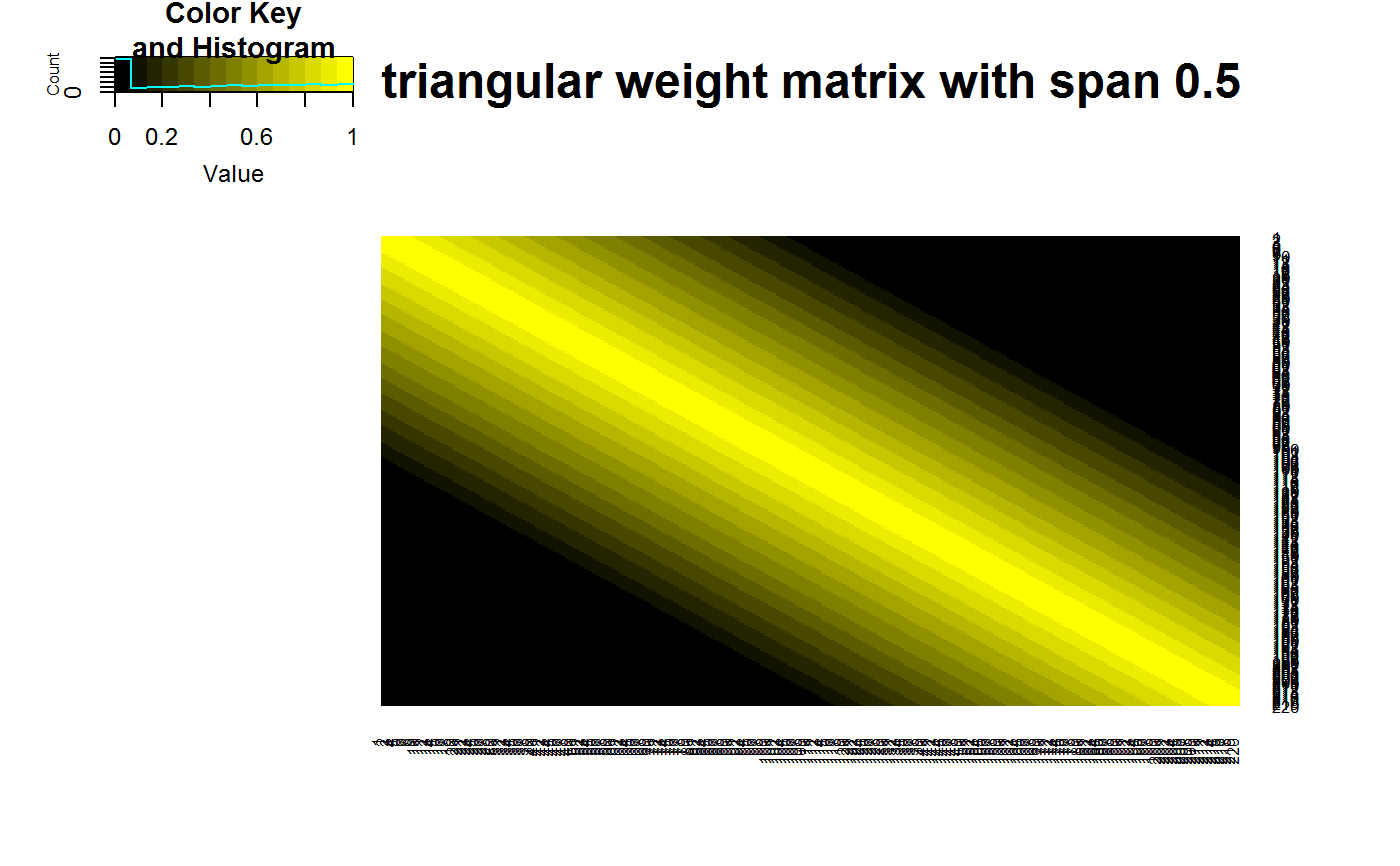
data(STRING) data(SKCM) SKCM_rank = t(apply(SKCM,1,rank)) # highly significantly DCARS gene pair: SKP1 and SKP2 # calculates p-value based on permutation DCARS(SKCM_rank,"SKP1","SKP2",plot=TRUE)#>#>#>#> [1] 0# extract only the test statistic DCARS(SKCM_rank,"SKP1","SKP2", extractTestStatisticOnly = TRUE)#>#>#> [1] 0.227379# not significantly DCARS gene pair: EIF3C and EIF5B # calculates p-value based on permutation DCARS(SKCM_rank,"EIF3C","EIF5B",plot=TRUE)#>#>#>#> [1] 0.97# extract only the test statistic DCARS(SKCM_rank,"EIF3C","EIF5B", extractTestStatisticOnly = TRUE)#>#>#> [1] 0.01790925# build weight matrix W = weightMatrix(ncol(SKCM_rank), type = "triangular", span = 0.5, plot = TRUE)#>#> #>#> #> #># extract DCARS test statistics SKCM_stats = DCARSacrossNetwork(SKCM_rank,edgelist = STRING, W = W, extractTestStatisticOnly = TRUE, verbose = FALSE) sort(SKCM_stats,decreasing=TRUE)[1:10]#> GMPS_IMPDH1 SKP1_SKP2 COPB2_COPE ANAPC10_ANAPC5 TBL3_UTP18 #> 0.2518867 0.2273790 0.2075771 0.1800155 0.1672255 #> PSMD12_PSMD3 POLR3B_POLR3D RAD18_UBE2B SNRPB_SNRPG RPL26L1_RPL8 #> 0.1659466 0.1629938 0.1621321 0.1555048 0.1531930