plotHigherOrderSequence
plotHigherOrderSequence.Rdthe plotHigherOrderSequence function plots weighted higher order statistic vectors (stored in higherOrderSequence) as line plots
plotHigherOrderSequence(
scHOT,
gene,
positionType = NULL,
branches = NULL,
positionColData = NULL
)Arguments
- scHOT
A scHOT object with higherOrderSequence in scHOT_output slot
- gene
is either a logical vector matching rows of entries in wcorsList, or a character of a gene
- positionType
A string indicates the position type, either trajectory or spatial
- branches
A character indicates that the colnames stored the branch information in colData (for trajectory type of data)
- positionColData
A vector indicates column names of colData that stored the postion informaton (for spatial type of data)
Value
ggplot object with line plots
Examples
data(liver)
scHOT_traj <- scHOT_buildFromMatrix(
mat = liver$liver_branch_hep,
cellData = list(pseudotime = liver$liver_pseudotime_hep),
positionType = "trajectory",
positionColData = "pseudotime")
scHOT_traj
#> class: scHOT
#> dim: 568 408
#> metadata(0):
#> assays(1): expression
#> rownames(568): 2810474O19Rik Abca1 ... Ahsg Epcam
#> rowData names(0):
#> colnames(408): E10.5D_3_02 E10.5D_2_01 ... E17.5D_1_01 F5A_E16.5
#> colData names(1): pseudotime
#> reducedDimNames(0):
#> mainExpName: NULL
#> altExpNames(0):
#> testingScaffold dim: 0 0
#> weightMatrix dim: 0 0
#> scHOT_output colnames (0):
#> param names (0):
#> position type: trajectory
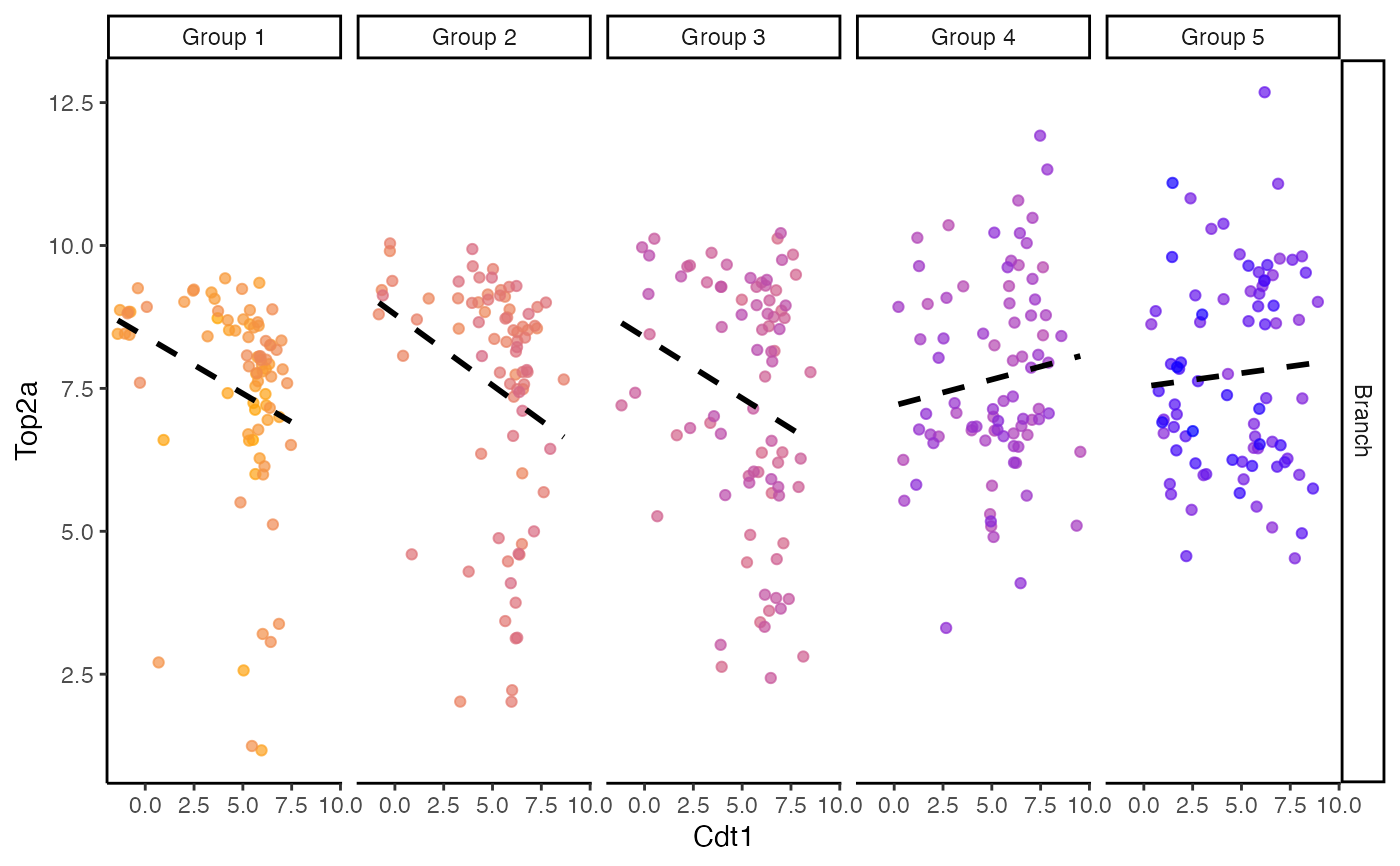
plotColouredExpression(scHOT_traj, c("Cdt1","Top2a"), n = 5)
#> ranked_by information is not provided,
#> the expression data is ranked by the branches
#> branches information is not provided
#> Warning: Use of `gdf_sub$ExpressionGene1` is discouraged. Use `ExpressionGene1` instead.
#> Warning: Use of `gdf_sub$ExpressionGene2` is discouraged. Use `ExpressionGene2` instead.
#> Warning: Use of `gdf_sub$ExpressionGene1` is discouraged. Use `ExpressionGene1` instead.
#> Warning: Use of `gdf_sub$ExpressionGene2` is discouraged. Use `ExpressionGene2` instead.
#> `geom_smooth()` using formula 'y ~ x'
 scHOT_traj <- scHOT_addTestingScaffold(scHOT_traj,
t(as.matrix(c("Cdt1", "Top2a"))))
scHOT_traj <- scHOT_setWeightMatrix(scHOT_traj,
positionColData = c("pseudotime"),
positionType = "trajectory",
nrow.out = NULL,
span = 0.25)
#> weightMatrix not provided, generating one using parameter settings...
#> type not specified, defaulting to triangular
scHOT_traj <- scHOT_calculateGlobalHigherOrderFunction(scHOT_traj,
higherOrderFunction =
weightedSpearman,
higherOrderFunctionType =
"weighted")
#> higherOrderFunctionType given will replace any stored param
#> higherOrderFunction given will replace any stored param
scHOT_traj <- scHOT_calculateHigherOrderTestStatistics(scHOT_traj,
higherOrderSummaryFunction =
sd)
#> higherOrderSummaryFunction will replace any stored param
slot(scHOT_traj, "scHOT_output")
#> DataFrame with 1 row and 5 columns
#> gene_1 gene_2 globalHigherOrderFunction
#> <character> <character> <numeric>
#> 1 Cdt1 Top2a -0.107678
#> higherOrderSequence higherOrderStatistic
#> <NumericList> <numeric>
#> 1 -0.475231,-0.474384,-0.471804,... 0.21339
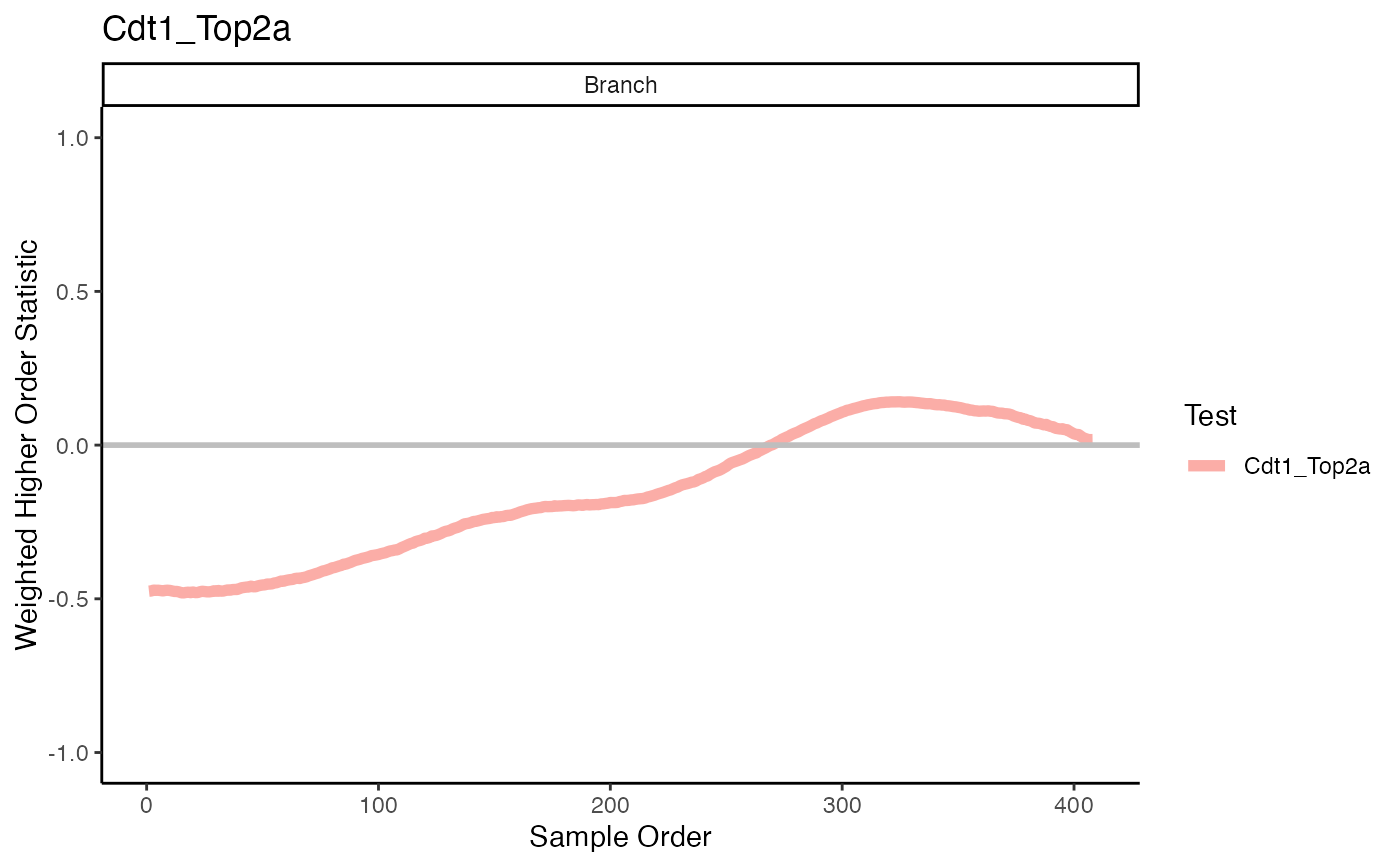
plotHigherOrderSequence(scHOT_traj, c("Cdt1_Top2a"))
#> branches information is not provided
#> Warning: 'as.is' should be specified by the caller; using TRUE
#> Warning: Use of `branch_long$SampleOrder` is discouraged. Use `SampleOrder` instead.
#> Warning: Use of `branch_long$WeightedCorrelation` is discouraged. Use `WeightedCorrelation` instead.
#> Warning: Use of `branch_long$GenePair` is discouraged. Use `GenePair` instead.
#> Warning: Use of `branch_long$GenePair` is discouraged. Use `GenePair` instead.
scHOT_traj <- scHOT_addTestingScaffold(scHOT_traj,
t(as.matrix(c("Cdt1", "Top2a"))))
scHOT_traj <- scHOT_setWeightMatrix(scHOT_traj,
positionColData = c("pseudotime"),
positionType = "trajectory",
nrow.out = NULL,
span = 0.25)
#> weightMatrix not provided, generating one using parameter settings...
#> type not specified, defaulting to triangular
scHOT_traj <- scHOT_calculateGlobalHigherOrderFunction(scHOT_traj,
higherOrderFunction =
weightedSpearman,
higherOrderFunctionType =
"weighted")
#> higherOrderFunctionType given will replace any stored param
#> higherOrderFunction given will replace any stored param
scHOT_traj <- scHOT_calculateHigherOrderTestStatistics(scHOT_traj,
higherOrderSummaryFunction =
sd)
#> higherOrderSummaryFunction will replace any stored param
slot(scHOT_traj, "scHOT_output")
#> DataFrame with 1 row and 5 columns
#> gene_1 gene_2 globalHigherOrderFunction
#> <character> <character> <numeric>
#> 1 Cdt1 Top2a -0.107678
#> higherOrderSequence higherOrderStatistic
#> <NumericList> <numeric>
#> 1 -0.475231,-0.474384,-0.471804,... 0.21339
plotHigherOrderSequence(scHOT_traj, c("Cdt1_Top2a"))
#> branches information is not provided
#> Warning: 'as.is' should be specified by the caller; using TRUE
#> Warning: Use of `branch_long$SampleOrder` is discouraged. Use `SampleOrder` instead.
#> Warning: Use of `branch_long$WeightedCorrelation` is discouraged. Use `WeightedCorrelation` instead.
#> Warning: Use of `branch_long$GenePair` is discouraged. Use `GenePair` instead.
#> Warning: Use of `branch_long$GenePair` is discouraged. Use `GenePair` instead.